In addition to the primary endpoint results from the CREATE trial, which you can read more about in detail here or as published in the New England Journal of Medicine, there was also a continuation phase study of the CREATE trial. This meant that all participants from the CREATE trial, including those who were randomized to the automated insulin delivery (AID) arm and those who were randomized to sensor-augmented insulin pump therapy (SAPT, which means just a pump and CGM, no algorithm), had the option to continue for another 24 weeks using the open source AID system.
These results were presented by Dr. Mercedes J. Burnside at #EASD2022, and I’ve summarized her presentation and the results below on behalf of the CREATE study team.
What is the “continuation phase”?
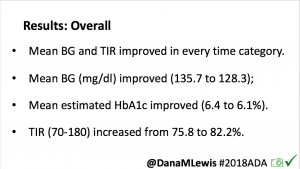
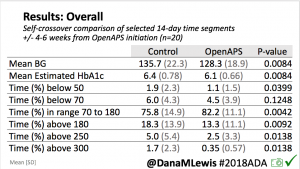
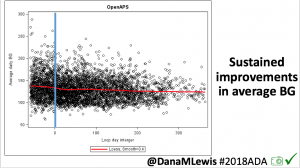
The CREATE trial was a multi-site, open-labeled, randomized, parallel-group, 24-week superiority trial evaluating the efficacy and safety of an open-source AID system using the OpenAPS algorithm in a modified version of AndroidAPS. Our study found that across children and adults, the percentage of time that the glucose level was in the target range of 3.9-10mmol/L [70-180mg/dL] was 14 percentage points higher among those who used the open-source AID system (95% confidence interval [CI], 9.2 to 18.8; P<0.001) compared to those who used sensor augmented pump therapy; a difference that corresponds to 3 hours 21 minutes more time spent in target range per day. The system did not contribute to any additional hypoglycemia. Glycemic improvements were evident within the first week and were maintained over the 24-week trial. This illustrates that all people with T1D, irrespective of their level of engagement with diabetes self-care and/or previous glycemic outcomes, stand to benefit from AID. This initial study concluded that open-source AID using the OpenAPS algorithm within a modified version of AndroidAPS, a widely used open-source AID solution, is efficacious and safe. These results were from the first 24-week phase when the two groups were randomized into SAPT and AID, accordingly.
The second 24-week phase is known as the “continuation phase” of the study.
There were 52 participants who were randomized into the SAPT group that chose to continue in the study and used AID for the 24 week continuation phase. We refer to those as the “SAPT-AID” group. There were 42 participants initially randomized into AID who continued to use AID for another 24 weeks (the AID-AID group).
One slight change to the continuation phase was that those in the SAPT-AID used a different insulin pump than the one used in the primary phase of the study (and 18/42 AID-AID participants also switched to this different pump during the continuation phase), but it was a similar Bluetooth-enabled pump that was interoperable with the AID system (app/algorithm) and CGM used in the primary outcome phase.
All 42 participants in AID-AID completed the continuation phase; 6 participants (out of 52) in the SAPT-AID group withdrew. One withdrew from infusion site issues; three with pump issues; and two who preferred SAPT.
What are the results from the continuation phase?
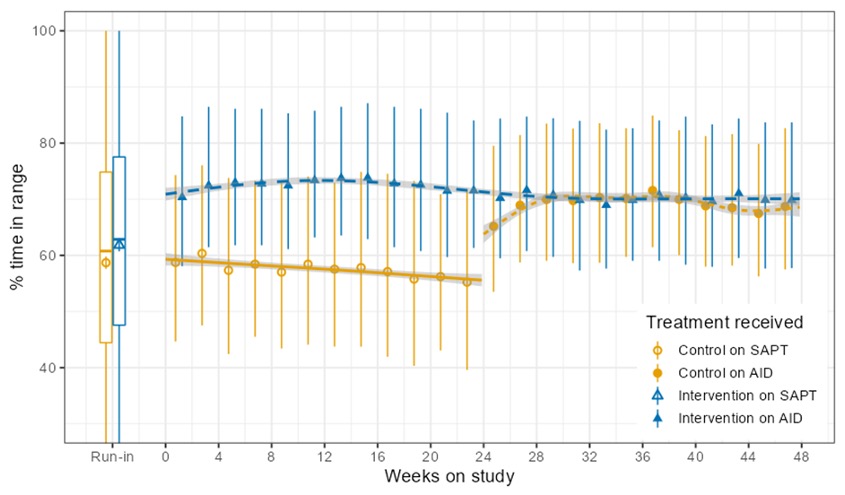
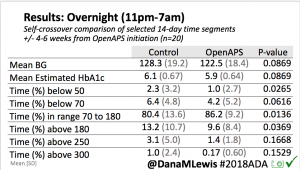
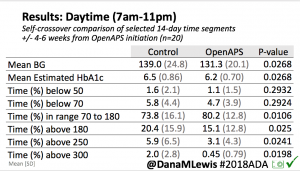
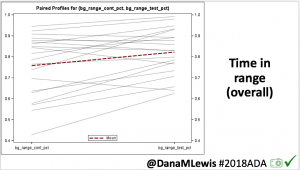
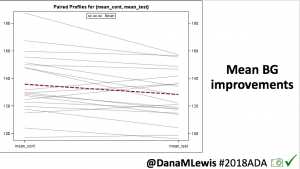
In the continuation phase, those in the SAPT-AID group saw a change in time in range (TIR) from 55±16% to 69±11% during the continuation phase when they used AID. In the SAPT-AID group, the percentage of participants who were able to achieve the target goals of TIR > 70% and time below range (TBR) <4% increased from 11% of participants during SAPT use to 49% during the 24 week AID use in the continuation phase. Like in the primary phase for AID-AID participants; the SAPT-AID participants saw the greatest treatment effect overnight with a TIR difference of 20.37% (95% CI, 17.68 to 23.07; p <0.001), and 9.21% during the day (95% CI, 7.44 to 10.98; p <0.001) during the continuation phase with open source AID.
Those in the AID-AID group, meaning those who continued for a second 24 week period using AID, saw similar TIR outcomes. Prior to AID use at the start of the study, TIR for that group was 61±14% and increased to 71±12% at the end of the primary outcome phase; after the next 6 months of the continuation phase, TIR was maintained at 70±12%. In this AID-AID group, the percentage of participants achieving target goals of TIR >70% and TBR <4% was 52% of participants in the first 6 months of AID use and 45% during the continuation phase. Similarly to the primary outcomes phase, in the continuation phase there was also no treatment effect by age interaction (p=0.39).
The TIR outcomes between both groups (SAPT-AID and AID-AID) were very similar after each group had used AID for 24 weeks (SAPT-AID group using AID for 24 weeks during the continuation phase and AID-AID using AID for 24 weeks during the initial RCT phase).. The adjusted difference in TIR between these groups was 1% (95% CI, -4 to 6; p=-0.67). There were no glycemic outcome differences between those using the two different study pumps (n=69, which was the SAPT-AID user group and 18 AID-AID participants who switched for continuation; and n=25, from the AID-AID group who elected to continue on the pump they used in the primary outcomes phase).
In the initial primary results (first 24 weeks of trial comparing the AID group to the SAPT group), there was a 14 percentage point difference between the groups. In the continuation phase, all used AID and the adjusted mean difference in TIR between AID and the initial SAPT results was a similar 12.10 percentage points (95% CI, p<0.001, SD 8.40).
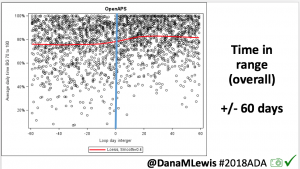
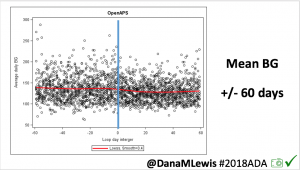
Similar to the primary phase, there was no DKA or severe hypoglycemia. Long-term use (over 48 weeks, representing 69 person-years) did not detect any rare severe adverse events.
Conclusion of the continuation study from the CREATE trial
In conclusion, the continuation study from the CREATE trial found that open-source AID using the OpenAPS algorithm within a modified version of AndroidAPS is efficacious and safe with various hardware (pumps), and demonstrates sustained glycaemic improvements without additional safety concerns.
Key points to takeaway:
- Over 48 weeks total of the study (6 months or 24 weeks in the primary phase; 6 months/24 weeks in the continuation phase), there were 64 person-years of use of open source AID in the study, compared to 59 person-years of use of sensor-augmented pump therapy.
- A variety of pump hardware options were used in the primary phase of the study among the SAPT group, due to hardware (pump) availability limitations. Different pumps were also used in the SAPT-AID group during the AID continuation phase, compared to the pumps available in the AID-AID group throughout both phases of trial. (Also, 18/42 of AID-AID participants chose to switch to the other pump type during the continuation phase).
- The similar TIR results (14 percentage points difference in primary and 12 percentage points difference in continuation phase between AID and SAPT groups) shows durability of the open source AID and algorithm used, regardless of pump hardware.
- The SAPT-AID group achieved similar TIR results at the end of their first 6 months of use of AID when compared to the AID-AID group at both their initial 6 months use and their total 12 months/48 weeks of use at the end of the continuation phase.
- The safety data showed no DKA or severe hypoglycemia in either the primary phase or the continuation phases.
- Glycemic improvements from this version of open source AID (the OpenAPS algorithm in a modified version of AndroidAPS) are not only immediate but also sustained, and do not increase safety concerns.



























Recent Comments