I tweeted last week+, “I just realized I’ve now gone about 3 weeks without meal bolusing.” That means just a meal announcement (i.e. carb entry estimate, a la 30 carbs or 60 carbs or whatever, based on my IFTTT buttons). No manual bolus.
I kind of keep waiting for the other shoe to drop, because it sounds to good to be true. I’m sure you’re skeptical reading this.
I bet she’s doing SOME bolus.
Well, she must not be eating any carbs.
She must be having worse outcomes, bad post-meal BGs, etc.
Nope, nope, and nope.
- While I started testing this new set of features with partial boluses and worked my way down (see more below on the testing topic), I’m now literally doing no manual meal bolus. I start eating, and press one button on my watch for a carb estimate entry (that via IFTTT goes to Nightscout and my rig).
- I eat carbs. I’ve eaten 120 grams of carbs of gluten free biscuits and gravy; 60-90 grams of pasta; dinner followed by a few gluten free cookies, etc.
- More nuanced details below, but:
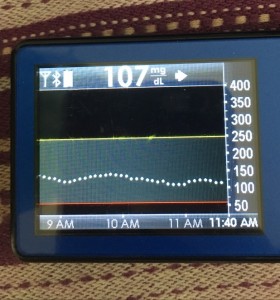
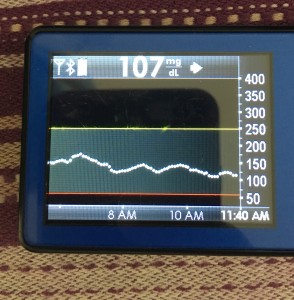
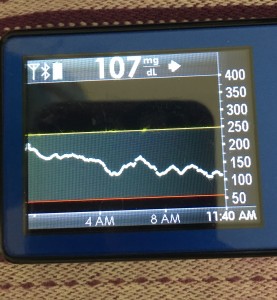
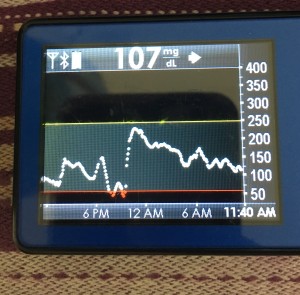
- My 70-180 time in range has stayed the same (93+%) compared to the versions I was testing before with manual meal boluses.
- My 70-150 and 80-160 time in ranges have decreased slightly compared to manual meal boluses, but…
- My average blood sugar has actually dropped down (as has my a1c to match).
- (So this means I’m having a few more spikes above 160, usually topping off in 160-170 whereas before my manual meal boluses would have me top off around 150, when all was well.)
Also note – no eating soon required. No early bolus or pre-bolus. Just single button press as I stick food in my mouth.
Wow.
(See where I said, waiting for the other shoe to drop?)
That’s why I waited a while to even tweet about it. Maybe it’s a fluke. Maybe it won’t work for other people. Maybe, maybe, maybe. Who knows. It’s still fairly early to tell, but as other people are beginning to test the current dev branch of oref0 with 0.6.0-related features, other people are starting to see improvements as well. (And that could be some of the many other features we are adding to 0.6.0, ranging from exponential curves for insulin activity, to allowing SMBs to do more, to carb-ratio-tuned-autosensitivity, to huge autotune improvements, etc.)
So while I don’t want to over-hype – and never do, what works for me will not work for everyone – I do want to share my cautious excitement over continuing to be able to push the envelope on algorithms and what might be possible outcome-wise for this kind of technology.
Here’s what is enabling me to be in the no-bolus zone for now well over a month, with still (to me) great outcomes worth the tradeoffs described above:
- Faster insulin. Thanks to our lovely looping friends in Germany/Austria, we came back from Europe with a few vials of Fiasp to try. I was HIGHLY skeptical about this. Some of our European friends saw great results right away, others didn’t. I didn’t get great results on it at first. Some of that may be due to natural changes between insulin types and not knowing exactly how to adjust my manual bolus strategy to the faster insulin action, but until we did some code changes to allow SMB‘s to do more and added some other features to what’s now 0.6.0, I wasn’t thrilled and in fact after about two weeks of it was about to switch off of it. So that brings me to #2.
- More improvements to the algorithm, which is now what will become the 0.6.0 release of oref0. There’s a whole lot of stuff packed in there. Exponential curves. Different carb absorption decay calculations. Allowing SMB to do more. Additional safety guards since we ramped SMB up.
How we started testing no-bolus approach:
- I have always known that about 6u of insulin (thanks to testing dating back to my early DIYPS days, many many many moons ago) is about as much as I should bolus at any time. So, even if I ate 120 carbs, I usually did about a 6u bolus up front, and let the rig pick up the rest as needed over more hours. I started doing ~75% or something like that of boluses, based on wherever I felt like rounding to with my easy bolus buttons.
- Whether I did 75% or 100%, I didn’t see a ton of difference at first…
- ..so I took a leap and tried no-bolus with some SMB adjustments to allow it to ramp up faster with carb entry. Behaviorally, I find it a lot easier to do nothing 😀 vs. figure out the right amount of up front bolus. And outcomes wise (see above) it was very similar.
It definitely was an interesting approach to test. Between the Fiasp and the no-bolus up front, in some meals it matched really well and I had practically no rise. Due to incoming netIOB, food type, etc, sometimes I did have a rise – but while it spiked slightly higher (160-170 usually vs my earlier 150s with manual bolus), it was only up there for 2-3 data points and then came sharply down, leveling out smoothly in my preferred post-meal range. So an important lesson I learned was not to over-react to just the BG curve going up, without looking at the predictions to see where I was going to come just back down. (And as I had more than one meal where the spike and drop back to normal happened, it was very easy to adjust to the BG graph and not get that emotional tug to “do more” with a quick short rise like that).
Obviously, starting BG makes a difference. I’m usually starting <130 mg/dL when I see these spikes cap out at 170 or lower. I’ve started higher, and seen higher rises, too. They’re not all perfect: with occasional pump site issues, carb underestimates, unplanned carb stacking, and all the randomness of diabetes and a non-structured lifestyle (including live-testing bleeding edge algorithm changes), I’ve spent 12% of the last month >160 mg/dL, which is about the same as the 3 months before that. But in most cases (I’d say 95%), the no-bolus approach has actually yielded better outcomes than I expected AND has avoided post-meal lows better than I would have achieved with a manual bolus.
This is huge when you think about the QOL aspect of not having to do as much math at a meal; and when you think about all the complicating factors related to food – timing (do you bolus when you order, or when the food arrives, or earlier than that?), and the gluten factor. I have celiac disease, so if I’m eating out (which we do a lot, and especially since I travel frequently), bolusing prior to setting eyes on the food (knowing they didn’t plate it with bread, causing them to have to go back and start all over again) just isn’t smart. That’s why eating soon historically worked so well for me vs. traditional pre-boluses, because I could set the target entering the restaurant, bolus when I laid eyes on my hopefully safe food, and get reasonable (150 topping out) meal outcomes.
It also worked really well in the case where a restaurant cooked my gluten free pasta in the same pasta cooker and water as regular pasta, but didn’t inform me until after I found stray gluten noodles in the bottom of my pasta dish and started asking how that was possible since they (used to) do gluten free well. (Now, I pick up heaps of pasta, and sort pasta noodles one by one to make sure they all match before ever eating gluten free pasta. It makes waiters look at you very worriedly as you wave pasta around in the air, but better safe than glutened (again).) So, I was majorly glutened, and my digestion system was all out of sorts (isn’t that a nice polite way to describe getting glutened?) for many days, which of course impacted BG and insulin right then and for the days afterward. But because I had done carb entry and no-bolus, I was able to edit the carb entry down, and I didn’t have that much insulin stacked, and didn’t end up low after glutening, which is usually what happens.
Is that a super regular situation for most people? No. But it was super nice. And also helped me face pasta again last night, so I could put in a (very low in case of gluten) carb estimate, match my noodles, eat pasta, and let the SMBs ramp up to match absorption. It works very well for me.
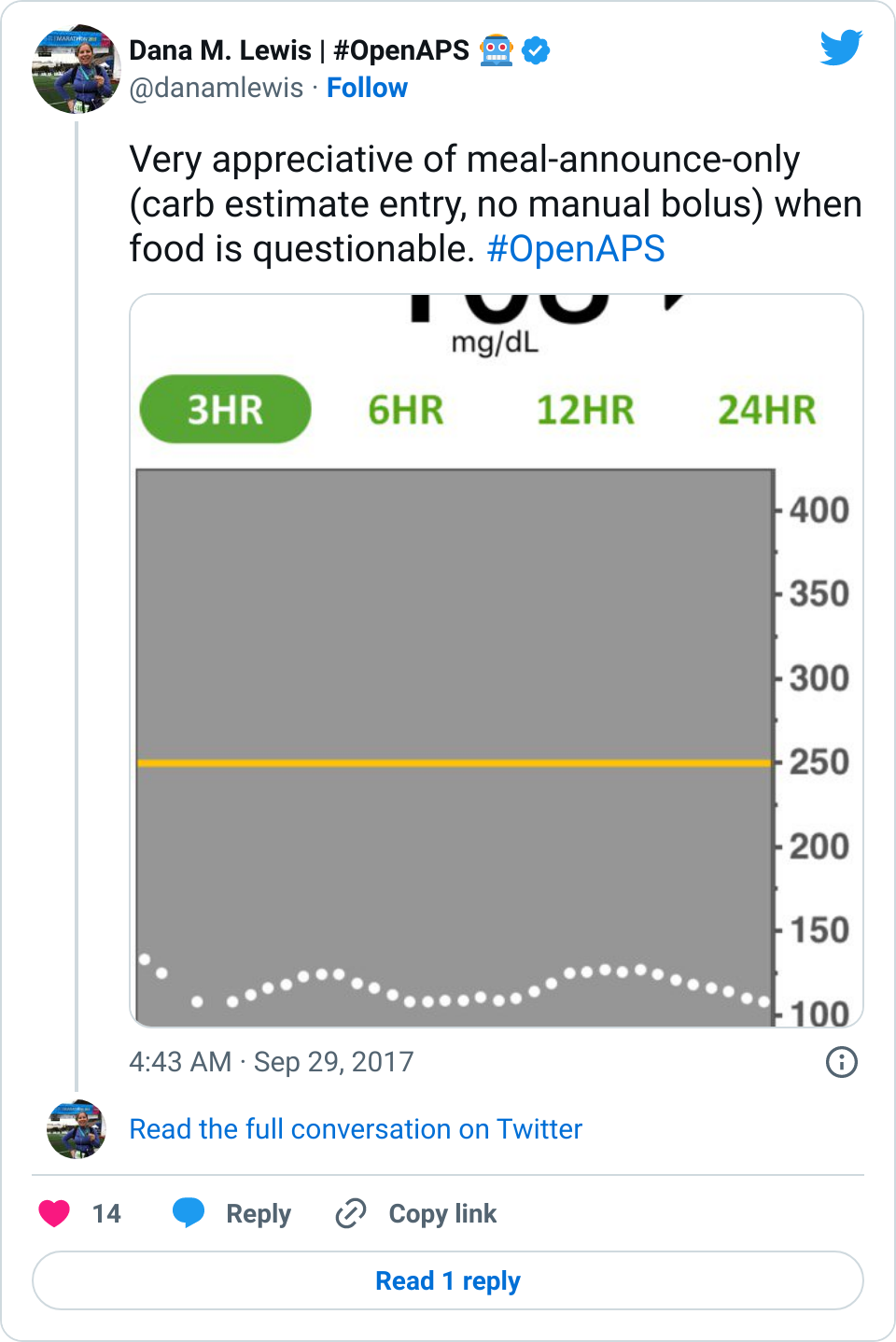
Whether you have celiac or not, for many reasons (insert yours here), it’s nice to not to have to commit to the bolus up front. It’s closer to approaching what I think non-PWDs do at mealtimes: just eat.
(I haven’t done much testing (yet? TBD) for no-carb-entry and no-meal-bolus scenario, I expect I would have higher spikes but would be interesting to see if it would still come down reasonably fast. Probably wouldn’t be my go-to strategy because I don’t mind a general meal size estimate one button push, but would be nice to know what that curve shape would look like. If I test that, it’ll start with small snacks and ramp my way up.)
The questions I always get:
- Q: HOW DO I GET THIS?
A: Caution: like all things OpenAPS but especially always true for the development branch, 0.6.0 is NOT released yet to master and is still highly experimental. I wouldn’t install dev unless you want to pay lots of close attention to it, and are willing to update multiple times over the course of the week, because Scott and I are merging features and tweaks almost daily to it.Got the disclaimers down? Ok. It’s in the dev branch of oref0. You should read this PR with notes on some more detail of what’s included, but you should also review the code diff to see all that’s changed, because it’s not all documented yet. Also, follow the instructions at the bottom to be able to install it without git. Hop into Gitter if you have questions about it!
(Big huge thanks to folks like Tim and Matthias for early testing of 0.6.0; and to Tim for writing up about the initial rounds of 0.6.0-dev here (note that we’ve made further changes since this post), and others who’ve been testing & providing feedback and input into the dev branch!)
- Q: When will this get “released” to master?
A: It depends. This is still a highly active dev branch, and we’re making a lot of changes and tweaking and testing things. The more people who test now and provide feedback will enable us to get to the final “prepare for release” testing stage. Lots and lots of testing, and things depend on how much existing needs tweaked, and what else we decide should go with this release. So, there’s never any specific release date. - Q: What is Fiasp?
A: Faster acting insulin that was only approved in Europe and Canada…until today. Convenient timing. I asked a PR person who messaged me about it, and they said it’s estimated to be available in U.S. pharmacies by late December/earlier Q1. As previously stated, available elsewhere in other parts of the world.Fiasp peaks sooner (say, ~45 minutes) with the same tail as everything else. It’s not instantaneous. For your million and one questions about whether it’s approved for your use in a tree, on a plane, at the zoo, and all other extrapolations – please ask Google/your doctor/the manufacturer, and not me. I don’t know.

- Q: Will any of this work for people NOT on Fiasp?
A: Nothing is guaranteed (even for other people on Fiasp), but the folks who’ve started testing 0.6.0 even without Fiasp (on Humalog or Novolog/Novorapid, etc.) have been happier on it vs. earlier versions, too.I don’t expect Fiasp to work super well forever for me, given what I’ve heard from other people with months of experience on it…and given my first two weeks of Fiasp not being spectacular, I want people to not expect miracles. (Sorry, this blog post does not promise miracles, so sorry if you got super excited at the above. No miracles! This is not a cure! We still have diabetes!) Like all things artificial pancreas, I think it’s better to be cautiously hopeful with realistic expectations that things *might* be a little bit better than before, but as always, YDMV (your diabetes may/will always vary), your body will vary, and life happens, etc. so who knows.
Just 4 months ago, we published a blog post pointing out that the new features had allowed us to achieve 4 out of 5 of: no bolus; not counting carbs, medium/high carb meals, 80%+ time in range; and no hypoglycemia. With Fiasp and 0.6.0 (currently what’s in the dev branch), we’ve now achieved all 5 simultaneously: I can eat large high-carb meals, enter very vague guesstimates of 60 or 90 carbs (no need for actual carb counting, just general size-based meal announcement), and still achieve 80%+ time in range 70-150 mg/dL without ever going <55 mg/dL. Does that mean that OpenAPS with Fiasp finally meets the definition of a “real” Artificial Pancreas (step 5 on JDRF’s 6-step AP development pathway)? We think it does.
So, tl;dr (because long post is long): with Fiasp and 0.6.0-dev branch, I’m able to not bolus for meals, and just enter a very generally sized meal estimate. It’s working well for me, and like all things, we’re working to make it available to other people via OpenAPS for others who want to try similar features/approaches. It may not work well for everyone. If it helps one other person, though, like everything else it’ll be worth it. Big thanks to Scott for LOTS of development in 0.6.0 and partnership in design of these features; too many people to name for testing and providing feedback and helping iterate on these features; and to the entire community for being awesome and helping us to continue to push the envelope on what might be possible for those of us with type 1 diabetes. 















Recent Comments