I’ve been puzzled when I see people online say that LLM’s “don’t write good code”. In my experience, they do. But given that most of these LLMs are used in chatbot mode – meaning you chat and give it instructions to generate the code – that might be where the disconnect lies. To get good code, you need effective prompting and to do so, you need clear thinking and ideas on what you are trying to achieve and how.
My recipe and understanding is:
Clear thinking + clear communication of ideas/request = effective prompting => effective code and other outputs
It also involves understanding what these systems can and can’t do. For example, as I’ve written about before, they can’t “know” things (although they can increasingly look things up) and they can’t do “mental” math. But, they can generally repeat patterns of words to help you see what is known about a topic and they can write code that you can execute (or it can execute, depending on settings) to solve a math problem.
What the system does well is help code small chunks, walk you through processes to link these sections of code up, and help you implement them (if you ask for it). The smaller the task (ask), the more effective it is. Or also – the easier it is for you to see when it completes the task and when it hasn’t been able to finish due to limitations like response length limits, information falling out of the context window (what it knows that you’ve told it); unclear prompting; and/or because you’re asking it to do things for which it doesn’t have expertise. Some of the last part – lack of expertise – can be improved with specific prompting techniques – and that’s also true for right-sizing the task it’s focusing on.
Right-size the task by giving a clear ask
If I were to ask an LLM to write me code for an iOS app to do XYZ, it could write me some code, but it certainly wouldn’t (at this point in history, written in February 2024), write all code and give me a downloadable file that includes it all and the ability to simply run it. What it can do is start writing chunks and snippets of code for bits and pieces of files that I can take and place and build upon.
How do I know this? Because I made that mistake when trying to build my first iOS apps in April and May 2023 (last year). It can’t do that (and still can’t today; I repeated the experiment). I had zero ideas how to build an iOS app; I had a sense that it involved XCode and pushing to the Apple iOS App Store, and that I needed “Swift” as the programming language. Luckily, though, I had a much stronger sense of how I wanted to structure the app user experience and what the app needed to do.
I followed the following steps:
- First, I initiated chat as a complete novice app builder. I told it I was new to building iOS apps and wanted to use XCode. I had XCode downloaded, but that was it. I told it to give me step by step instructions for opening XCode and setting up a project. Success! That was effective.
- I opened a different chat window after that, to start a new chat. I told it that it was an expert in iOS programming using Swift and XCode. Then I described the app that I wanted to build, said where I was in the process (e.g. had opened and started a project in XCode but had no code yet), and asked it for code to put on the home screen so I could build and open the app and it would have content on the home screen. Success!
- From there, I was able to stay in the same chat window and ask it for pieces at a time. I wanted to have a new user complete an onboarding flow the very first time they opened the app. I explained the number of screens and content I wanted on those screens; the chat was able to generate code, tell me how to create that in a file, and how to write code that would trigger this only for new users. Success!
- I was able to then add buttons to the home screen; have those buttons open new screens of the app; add navigation back to the home; etc. Success!
- (Rinse and repeat, continuing until all of the functionality was built out a step at a time).
To someone with familiarity building and programming things, this probably follows a logical process of how you might build apps. If you’ve built iOS apps before and are an expert in Swift programming, you’re either not reading this blog post or are thinking I (the human) am dumb and inexperienced.
Inexperienced, yes, I was (in April 2023). But what I am trying to show here is for someone new to a process and language, this is how we need to break down steps and work with LLMs to give it small tasks to help us understand and implement the code it produces before moving forward with a new task (ask). It takes these small building block tasks in order to build up to a complete app with all the functionality that we want. Nowadays, even though I can now whip up a prototype project and iOS app and deploy it to my phone within an hour (by working with an LLM as described above, but skipping some of the introductory set-up steps now that I have experience in those), I still follow the same general process to give the LLM the big picture and efficiently ask it to code pieces of the puzzle I want to create.
As the human, you need to be able to keep the big picture – full app purpose and functionality – in mind while subcontracting with the LLM to generate code for specific chunks of code to help achieve new functionality in our project.
In my experience, this is very much like pair programming with a human. In fact, this is exactly what we did when we built DIYPS over ten years ago (wow) and then OpenAPS within the following year. I’ve talked endlessly about how Scott and I would discuss an idea and agree on the big picture task; then I would direct sub-tasks and asks that he, then also Ben and others would be coding on (at first, because I didn’t have as much experience coding and this was 10 years ago without LLMs; I gradually took on more of those coding steps and roles as well). I was in charge of the big picture project and process and end goal; it didn’t matter who wrote which code or how; we worked together to achieve the intended end result. (And it worked amazingly well; here I am 10 years later still using DIYPS and OpenAPS; and tens of thousands of people globally are all using open source AID systems spun off of the algorithm we built through this process!)
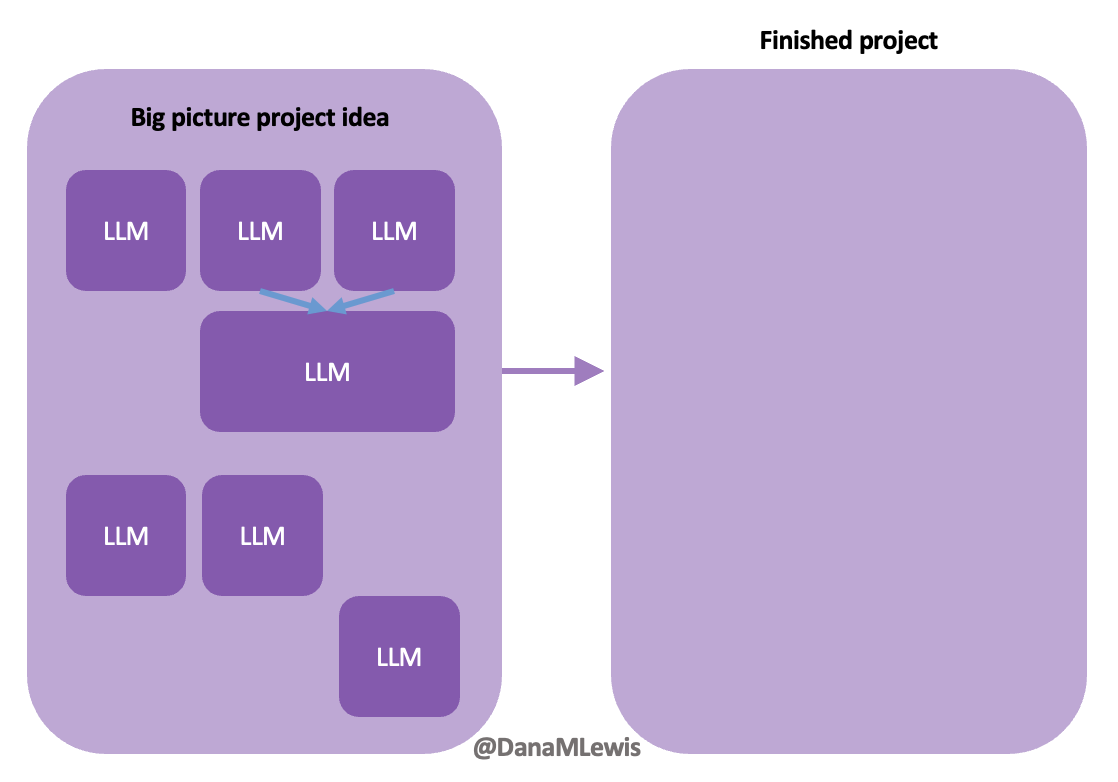 Today, I would say the same is true. It doesn’t matter – for my types of projects – if a human or an LLM “wrote” the code. What matters is: does it work as intended? Does it achieve the goal? Does it contribute to the goal of the project?
Today, I would say the same is true. It doesn’t matter – for my types of projects – if a human or an LLM “wrote” the code. What matters is: does it work as intended? Does it achieve the goal? Does it contribute to the goal of the project?
Coding can be done – often by anyone (human with relevant coding expertise) or anything (LLM with effective prompting) – for any purpose. The critical key is knowing what the purpose is of the project and keeping the coding heading in the direction of serving that purpose.
Tips for right-sizing the ask
- Consider using different chat windows for different purposes, rather than trying to do it all in one. Yes, context windows are getting bigger, but you’ll still likely benefit from giving different prompts in different windows (more on effective prompting below).Start with one window for getting started with setting up a project (e.g. how to get XCode on a Mac and start a project; what file structure to use for an app/project that will do XYZ; how to start a Jupyter notebook for doing data science with python; etc); brainstorming ideas to scope your project; then separately for starting a series of coding sub-tasks (e.g. write code for the home page screen for your app; add a button that allows voice entry functionality; add in HealthKit permission functionality; etc.) that serves the big picture goal.
- Make a list for yourself of the steps needed to build a new piece of functionality for your project. If you know what the steps are, you can specifically ask the LLM for that.Again, use a separate window if you need to. For example, if you want to add in the ability to save data to HealthKit from your app, you may start a new chat window that asks the LLM generally how does one add HealthKit functionality for an app? It’ll describe the process of certain settings that need to be done in XCode for the project; adding code that prompts the user with correct permissions; and then code that actually does the saving/revising to HealthKit.
Make your list (by yourself or with help), then you can go ask the LLM to do those things in your coding/task window for your specific project. You can go set the settings in XCode yourself, and skip to asking it for the task you need it to do, e.g. “write code to prompt the user with HealthKit permissions when button X is clicked”.
(Sure, you can do the ask for help in outlining steps in the same window that you’ve been prompting for coding sub-tasks, just be aware that the more you do this, the more quickly you’ll burn through your context window. Sometimes that’s ok, and you’ll get a feel for when to do a separate window with the more experience you get.)
- Pay attention as you go and see how much code it can generate and when it falls short of an ask. This will help you improve the rate at which you successfully ask and it fully completes a task for future asks. I observe that when I don’t know – due to my lack of expertise – the right size of a task, it’s more prone to give me ½-⅔ of the code and solution but need additional prompting after that. Sometimes I ask it to continue where it cut off; other times I start implementing/working with the bits of code (the first ⅔) it gave me, and have a mental or written note that this did not completely generate all steps/code for the functionality and to come back.Part of why sometimes it is effective to get started with ⅔ of the code is because you’ll likely need to debug/test the first bit of code, anyway. Sometimes when you paste in code it’s using methods that don’t match the version you’re targeting (e.g. functionality that is outdated as of iOS 15, for example, when you’re targeting iOS 17 and newer) and it’ll flag a warning or block it from working until you fix it.
Once you’ve debugged/tested as much as you can of the original ⅔ of code it gave you, you can prompt it to say “Ok, I’ve done X and Y. We were trying to (repeat initial instructions/prompt) – what are the remaining next steps? Please code that.” to go back and finish the remaining pieces of that functionality.
(Note that saying “please code that” isn’t necessarily good prompt technique, see below).
Again, much of this is paying attention to how the sub-task is getting done in service of the overall big picture goal of your project; or the chunk that you’ve been working on if you’re building new functionality. Keeping track with whatever method you prefer – in your head, a physical written list, a checklist digitally, or notes showing what you’ve done/not done – is helpful.
Most of the above I used for coding examples, but I follow the same general process when writing research papers, blog posts, research protocols, etc. My point is that this works for all types of projects that you’d work on with an LLM, whether the output generation intended is code or human-focused language that you’d write or speak.
But, coding or writing language, the other thing that makes a difference in addition to right-sizing the task is effective prompting. I’ve intuitively noticed that has made the biggest difference in my projects for getting the output matching my expertise. Conversely, I have actually peer reviewed papers for medical journals that do a horrifying job with prompting. You’ll hear people talk about “prompt engineering” and this is what it is referring to: how do you engineer (write) a prompt to get the ideal response from the LLM?
Tips for effective prompting with an LLM
- Personas and roles can make a difference, both for you and for the LLM. What do I mean by this? Start your prompt by telling the LLM what perspective you want it to take. Without it, you’re going to make it guess what information and style of response you’re looking for. Here’s an example: if you asked it what caused cancer, it’s going to default to safety and give you a general public answer about causes of cancer in very plain, lay language. Which may be fine. But if you’re looking to generate a better understanding of the causal mechanism of cancer; what is known; and what is not known, you will get better results if you prompt it with “You are an experienced medical oncologist” so it speaks from the generated perspective of that role. Similarly, you can tell it your role. Follow it with “Please describe the causal mechanisms of cancer and what is known and not known” and/or “I am also an experienced medical researcher, although not an oncologist” to help contextualize that you want a deeper, technical approach to the answer and not high level plain language in the response.
Compare and contrast when you prompt the following:
A. “What causes cancer?”
B. “You are an experienced medical oncologist. What causes cancer? How would you explain this differently in lay language to a patient, and how would you explain this to another doctor who is not an oncologist?”
C. “You are an experienced medical oncologist. Please describe the causal mechanisms of cancer and what is known and not known. I am also an experienced medical researcher, although not an oncologist.”
You’ll likely get different types of answers, with some overlap between A and the first part of answer B. Ditto for a tiny bit of overlap between the latter half of answer B and for C.
I do the same kind of prompting with technical projects where I want code. Often, I will say “You are an expert data scientist with experience writing code in Python for a Jupyter Notebook” or “You are an AI programming assistant with expertise in building iOS apps using XCode and SwiftUI”. Those will then be followed with a brief description of my project (more on why this is brief below) and the first task I’m giving it.
The same also goes for writing-related tasks; the persona I give it and/or the role I reference for myself makes a sizable difference in getting the quality of the output to match the style and quality I was seeking in a response.
- Be specific. Saying “please code that” or “please write that” might work, sometimes, but more often or not will get a less effective output than if you provide a more specific prompt.I am a literal person, so this is something I think about a lot because I’m always parsing and mentally reviewing what people say to me because my instinct is to take their words literally and I have to think through the likelihood that those words were intended literally or if there is context that should be used to filter those words to be less literal. Sometimes, you’ll be thinking about something and start talking to someone about something, and they have no idea what on earth you’re talking about because the last part of your out-loud conversation with them was about a completely different topic!
LLMs are the same as the confused conversational partner who doesn’t know what you’re thinking about. LLMs only know what you’ve last/recently told it (and more quickly than humans will ‘forget’ what you told it about a project). Remember the above tips about brainstorming and making a list of tasks for a project? Providing a description of the task along with the ask (e.g. we are doing X related to the purpose of achieving Y, please code X) will get you better output more closely matching what you wanted than saying “please code that” where the LLM might code something else to achieve Y if you didn’t tell it you wanted to focus on X.
I find this even more necessary with writing related projects. I often find I need to give it the persona “You are an expert medical researcher”, the project “we are writing a research paper for a medical journal”, the task “we need to write the methods section of the paper”, and a clear ask “please review the code and analyses and make an outline of the steps that we have completed in this process, with sufficient detail that we could later write a methods section of a research paper”. A follow up ask is then “please take this list and draft it into the methods section”. That process with all of that specific context gives better results than “write a methods section” or “write the methods” etc.
- Be willing to start over with a new window/chat. Sometimes the LLM can get itself lost in solving a sub-task and lose sight (via lost context window) of the big picture of a project, and you’ll find yourself having to repeat over and over again what you’re asking it to do. Don’t be afraid to cut your losses and start a new chat for a sub-task that you’ve been stuck on. You may be able to eventually come back to the same window as before, or the new window might become your new ‘home’ for the project…or sometimes a third, fourth, or fifth window will.
- Try, try again.
I may hold the record for the longest running bug that I (and the LLM) could. Not. solve. This was so, so annoying. No users apparently noticed it but I knew about it and it bugged me for months and months. Every few weeks I would go to an old window and also start a new window, describe the problem, paste the code in, and ask for help to solve it. I asked it to identify problems with the code; I asked it to explain the code and unexpected/unintended functionality from it; I asked it what types of general things would be likely to cause that type of bug. It couldn’t find the problem. I couldn’t find the problem. Finally, one day, I did all of the above, but then also started pasting every single file from my project and asking if it was likely to include code that could be related to the problem. By forcing myself to review all my code files with this problem in mind, even though the files weren’t related at all to the file/bug….I finally spotted the problem myself. I pasted the code in, asked if it was a possibility that it was related to the problem, the LLM said yes, I tried a change and…voila! Bug solved on January 16 after plaguing me since November 8. (And probably existed before then but I didn’t have functionality built until November 8 where I realized it was a problem). I was beating myself up about it and posted to Twitter about finally solving the bug (but very much with the mindset of feeling very stupid about it). Someone replied and said “congrats! sounds like it was a tough one!”. Which I realized was a very kind framing and one that I liked, because it was a tough one; and also I am doing a tough thing that no one else is doing and I would not have been willing to try to do without an LLM to support.Similarly, just this last week on Tuesday I spent about 3 hours working on a sub-task for a new project. It took 3 hours to do something that on a previous project took me about 40 minutes, so I was hyper aware of the time mismatch and perceiving that 3 hours was a long time to spend on the task. I vented to Scott quite a bit on Tuesday night, and he reminded me that sure it took “3 hours” but I did something in 3 hours that would take 3 years otherwise because no one else would do (or is doing) the project that I’m working on. Then on Wednesday, I spent an hour doing another part of the project and Thursday whipped through another hour and a half of doing huge chunks of work that ended up being highly efficient and much faster than they would have been, in part because the “three hours” it took on Tuesday wasn’t just about the code but about organizing my thinking, scoping the project and research protocol, etc. and doing a huge portion of other work to organize my thinking to be able to effectively prompt the LLM to do the sub-task (that probably did actually take closer to the ~40 minutes, similar to the prior project).
All this to say: LLMs have become pair programmers and collaborators and writers that are helping me achieve tasks and projects that no one else in the world is working on yet. (It reminds me very much of my early work with DIYPS and OpenAPS where we did the work, quietly, and people eventually took notice and paid attention, albeit slower than we wished but years faster than had we not done that work. I’m doing the same thing in a new field/project space now.) Sometimes, the first attempt to delegate a sub-task doesn’t work. It may be because I haven’t organized my thinking enough, and the lack of ideal output shows that I have not prompted effectively yet. Sometimes I can quickly fix the prompt to be effective; but sometimes it highlights that my thinking is not yet clear; my ability to communicate the project/task/big picture is not yet sufficient; and the process of achieving the clarity of thinking and translating to the LLM takes time (e.g. “that took 3 hours when it should have taken 40 minutes”) but ultimately still moves me forward to solving the problem or achieving the tasks and sub-tasks that I wanted to do. Remember what I said at the beginning:
Clear thinking + clear communication of ideas/request = effective prompting => effective code and other outputs
- Try it anyway.
I am trying to get out of the habit of saying “I can’t do X”, like “I can’t code/program an iOS app”…because now I can. I’ve in fact built and shipped/launched/made available multiple iOS apps (check out Carb Pilot if you’re interested in macronutrient estimates for any reason; you can customize so you only see the one(s) you care about; or if you have EPI, check out PERT Pilot, which is the world’s first and only app for tracking pancreatic enzyme replacement therapy and has the same AI feature for generating macronutrient estimates to aid in adjusting enzyme dosing for EPI.) I’ve also made really cool, 100% custom-to-me niche apps to serve a personal purpose that save me tons of time and energy. I can do those things, because I tried. I flopped a bunch along the way – it took me several hours to solve a simple iOS programming error related to home screen navigation in my first few apps – but in the process I learned how to do those things and now I can build apps. I’ve coded and developed for OpenAPS and other open source projects, including a tool for data conversion that no one else in the world had built. Yet, my brain still tries to tell me I can’t code/program/etc (and to be fair, humans try to tell me that sometimes, too).I bring that up to contextualize that I’m working on – and I wish others would work on to – trying to address the reflexive thoughts of what we can and can’t do, based on prior knowledge. The world is different now and tools like LLMs make it possible to learn new things and build new projects that maybe we didn’t have time/energy to do before (not that we couldn’t). The bar to entry and the bar to starting and trying is so much lower than it was even a year ago. It really comes down to willingness to try and see, which I recognize is hard: I have those thought patterns too of “I can’t do X”, but I’m trying to notice when I have those patterns; shift my thinking to “I used to not be able to do X; I wonder if it is possible to work with an LLM to do part of X or learn how to do Y so that I could try to do X”.
A recent real example for me is power calculations and sample size estimates for future clinical trials. That’s something I can’t do; it requires a statistician and specialized software and expertise.
Or…does it?
I asked my LLM how power calculations are done. It explained. I asked if it was possible to do it using Python code in a Jupyter notebook. I asked what information would be needed to do so. It walked me through the decisions I needed to make about power and significance, and highlighted variables I needed to define/collect to put into the calculation. I had generated the data from a previous study so I had all the pieces (variables) I needed. I asked it to write code for me to run in a Jupyter notebook, and it did. I tweaked the code, input my variables, ran it..and got the result. I had run a power calculation! (Shocked face here). But then I got imposter syndrome again, reached out to a statistician who I had previously worked with on a research project. I shared my code and asked if that was the correct or an acceptable approach and if I was interpreting it correctly. His response? It was correct, and “I couldn’t have done it better myself”.
(I’m still shocked about this).
He also kindly took my variables and put it in the specialized software he uses and confirmed that the results output matched what my code did, then pointed out something that taught me something for future projects that might be different (where the data is/isn’t normally distributed) although it didn’t influence the output of my calculation for this project.
What I learned from this was a) this statistician is amazing (which I already knew from working with him in the past) and kind to support my learning like this; b) I can do pieces of projects that I previously thought were far beyond my expertise; c) the blocker is truly in my head, and the more we break out of or identify the patterns stopping us from trying, the farther we will get.
“Try it anyway” also refers to trying things over time. The LLMs are improving every few months and often have new capabilities that didn’t before. Much of my work is done with GPT-4 and the more nuanced, advanced technical tasks are way more efficient than when using GPT-3.5. That being said, some tasks can absolutely be done with GPT-3.5-level AI. Doing something now and not quite figuring it out could be something that you sort out in a few weeks/months (see above about my 3 month bug); it could be something that is easier to do once you advance your thinking ; or it could be more efficiently done with the next model of the LLM you’re working with.
- Test whether custom instructions help. Be aware though that sometimes too many instructions can conflict and also take up some of your context window. Plus if you forget what instructions you gave it, you might get seemingly unexpected responses in future chats. (You can always change the custom instructions and/or turn it on and off.)
I’m hoping this helps give people confidence or context to try things with LLMs that they were not willing to try before; or to help get in the habit of remembering to try things with LLMs; and to get the best possible output for the project that they’re working on.
Remember:
- Right-size the task by making a clear ask.
- You can use different chat windows for different levels of the same project.
- Use a list to help you, the human, keep track of all the pieces that contribute to the bigger picture of the project.
- Try giving the LLM a persona for an ask; and test whether you also need to assign yourself a persona or not for a particular type of request.
- Be specific, think of the LLM as a conversational partner that can’t read your mind.
- Don’t be afraid to start over with a new context window/chat.
- Things that were hard a year ago might be easier with an LLM; you should try again.
- You can do more, partnering with an LLM, than you can on your own, and likely can do things you didn’t realize were possible for you to do!
Clear thinking + clear communication of ideas/request = effective prompting => effective code and other outputs
—
Have any tips to help others get more effective output from LLMs? I’d love to hear them, please comment below and share your tips as well!






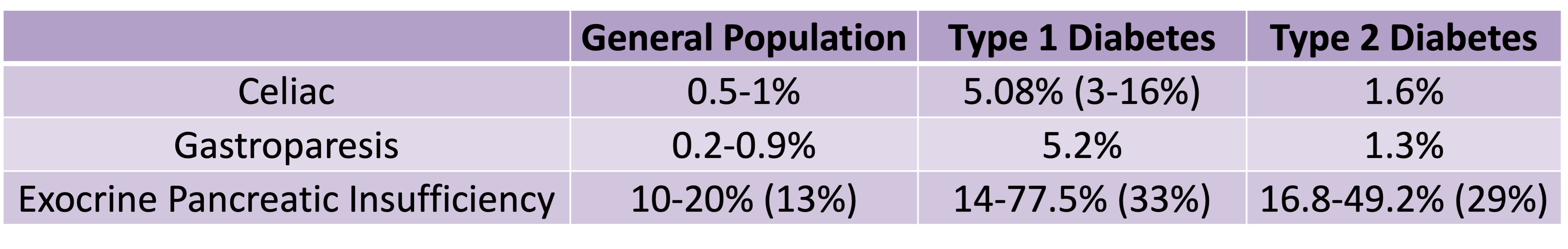 Celiac disease is more common in people with diabetes (~5%) than in the general public (0.5-1%). Gastroparesis, when gastric emptying is delayed, is also more common in people with diabetes (5% in PWD).However, the prevalence of EPI is 14-77.5% (median 33%) in Type 1 diabetes and 16.8-49.2% (median 29%) in Type 2 diabetes (and 5.4-77% prevalence when type of diabetes was not specified). This again is higher than general population prevalence of EPI.
Celiac disease is more common in people with diabetes (~5%) than in the general public (0.5-1%). Gastroparesis, when gastric emptying is delayed, is also more common in people with diabetes (5% in PWD).However, the prevalence of EPI is 14-77.5% (median 33%) in Type 1 diabetes and 16.8-49.2% (median 29%) in Type 2 diabetes (and 5.4-77% prevalence when type of diabetes was not specified). This again is higher than general population prevalence of EPI.


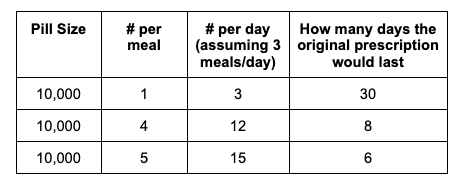
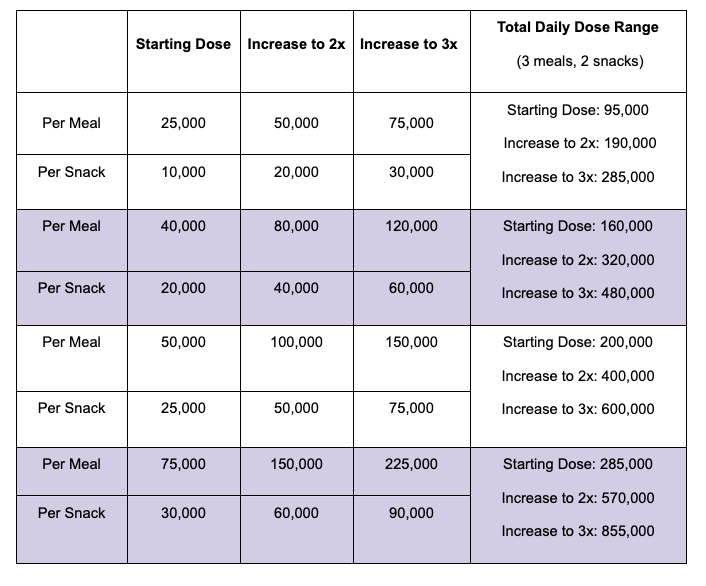
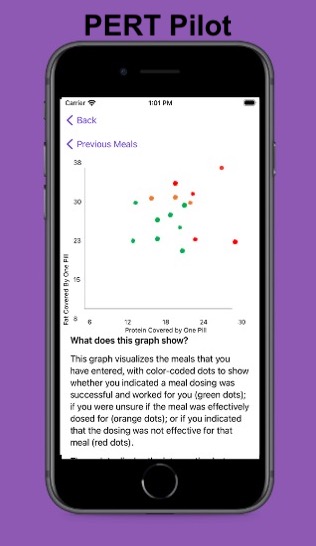 For example, I was initially prescribed an enzyme dose that was one pill per meal (and no snacks), so I had 3 pills per day. But I quickly found myself needing two pills per meal, based on what I was typically eating. I summarized my data to my doctor, saying that I found one pill typically covered up to ~30 grams of fat per meal, but most of my meals were >30 grams of fat, so that I wanted to update my prescription to have an average of 2 pills per meal of this prescription size. I also wanted to be able to eat snacks, so I asked for 2 pills per meal, 1 per snack, which meant that my prescription increased to 8 pills per day (of the same size), to cover 2 pills x 3 meals a day (=6) plus up to 2 snacks (=2). I also had weeks of data to show that my average meal was >30 grams of fat to confirm that I need more than the amount of lipase I was originally prescribed. My doctor was happy to increase my prescription as a result, and this is what I’ve been using successfully for over a year ever since.
For example, I was initially prescribed an enzyme dose that was one pill per meal (and no snacks), so I had 3 pills per day. But I quickly found myself needing two pills per meal, based on what I was typically eating. I summarized my data to my doctor, saying that I found one pill typically covered up to ~30 grams of fat per meal, but most of my meals were >30 grams of fat, so that I wanted to update my prescription to have an average of 2 pills per meal of this prescription size. I also wanted to be able to eat snacks, so I asked for 2 pills per meal, 1 per snack, which meant that my prescription increased to 8 pills per day (of the same size), to cover 2 pills x 3 meals a day (=6) plus up to 2 snacks (=2). I also had weeks of data to show that my average meal was >30 grams of fat to confirm that I need more than the amount of lipase I was originally prescribed. My doctor was happy to increase my prescription as a result, and this is what I’ve been using successfully for over a year ever since.

Recent Comments