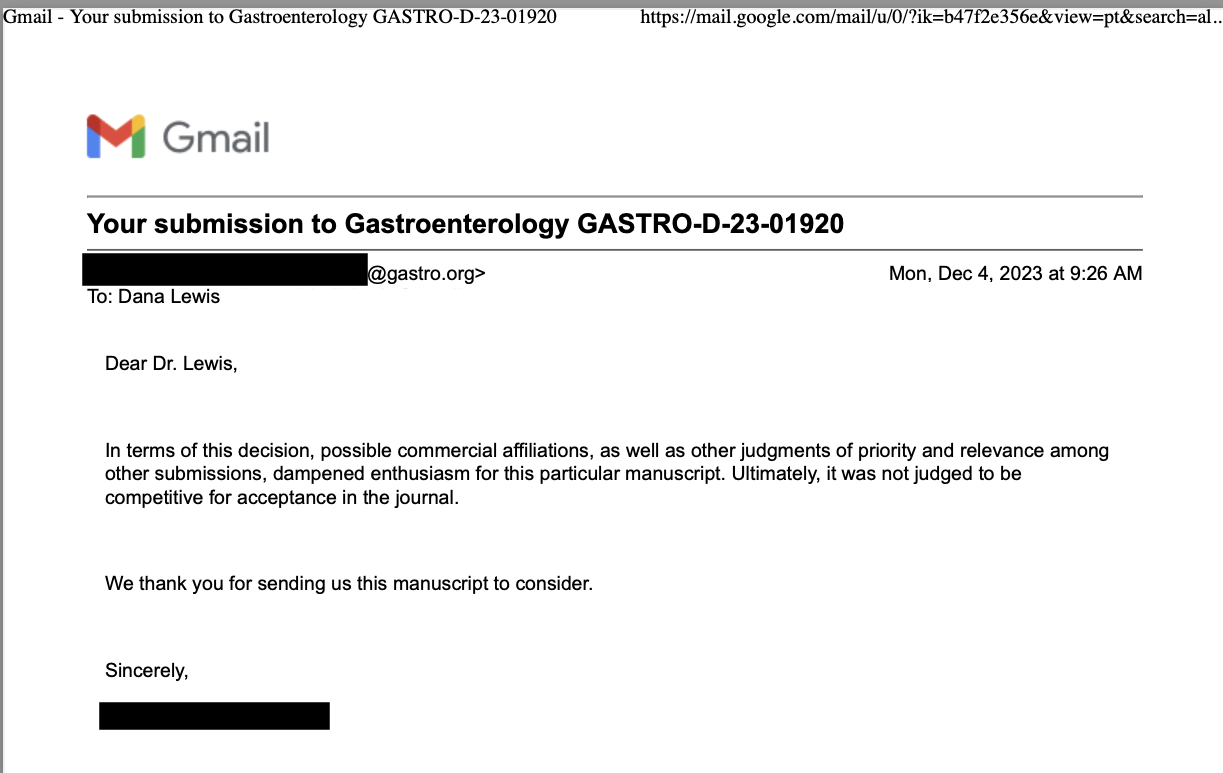
I was asked to contribute a talk in a session at Stanford’s recent AI+Health conference. I spoke alongside two clinician researchers, and we organized the session so that I would talk about some individual patient experiences & perspectives on AI use in health, then scale up to talk about health system use, then global perspectives on AI for health-related use cases.
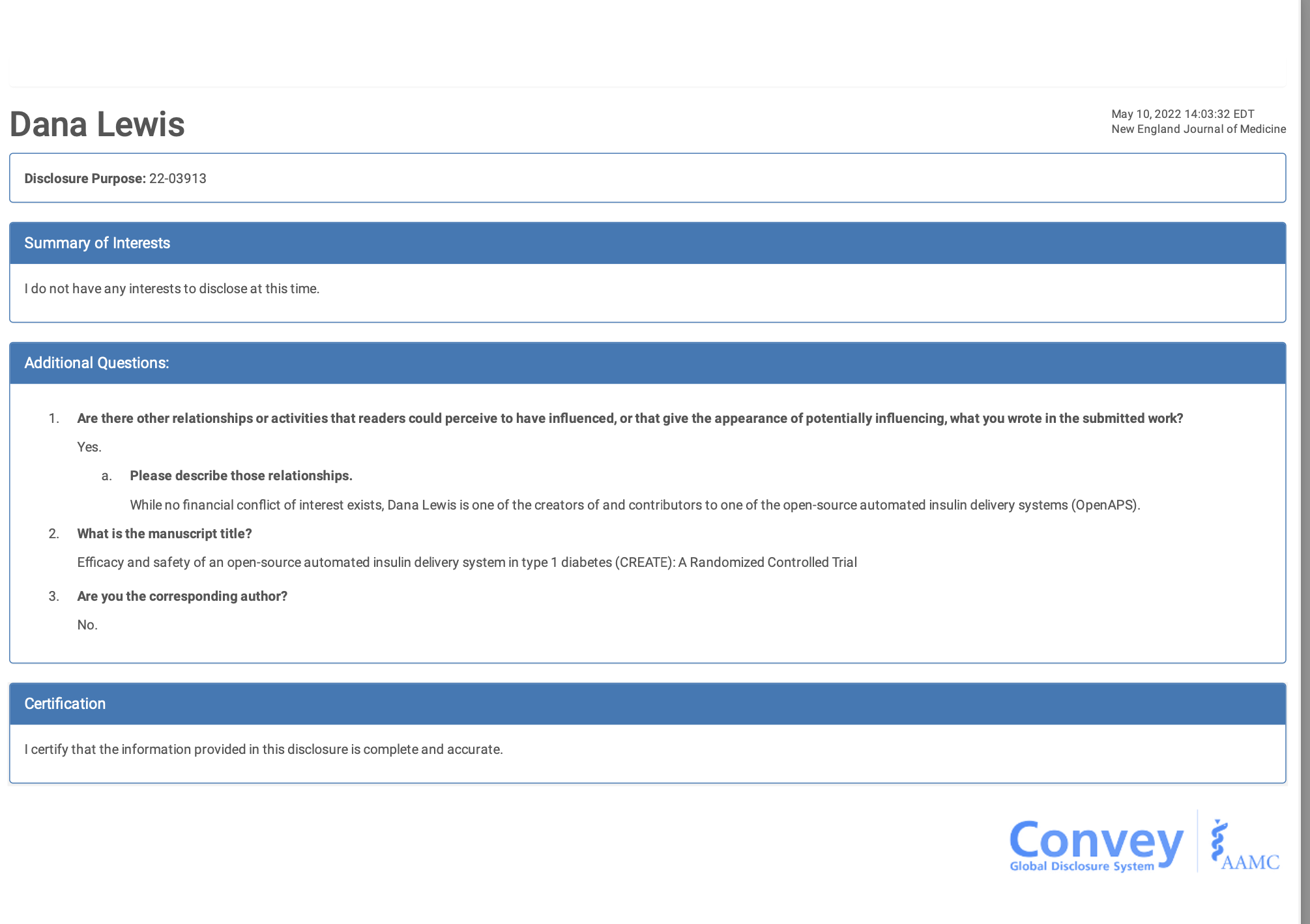
One thing I’ve been speaking about lately (including when I was asked to present in DC at the FDA AI workshop) is about how AI…is not one thing. There are different technologies, different models, and they’re going to work differently based on the prompts (like our research showed here about chart notes + LLM responses, as evaluated by a patient and clinician) as well as whether it’s a one-off use, a recurring use, something that’s well-defined in the literature and training materials or whether it’s not well-defined. I find myself stumbling into the latter areas quite a bit, and have a knack of finding ways to deal with this stuff, so I’m probably more attuned to these spaces than most: everything from figuring out how we people with diabetes can automate our own insulin delivery (because it was not commercially available for years); to figuring out how to titrate my own pancreatic enzyme replacement therapy and building an app to help others track and figure things out themselves; to more recent experiences with titration and finding a mechanistic model for an undiagnosed disease. The grey area, the bleeding edge, no-(wo)man’s land where no one has answers or ideas of what to do next…I hang out there a lot.
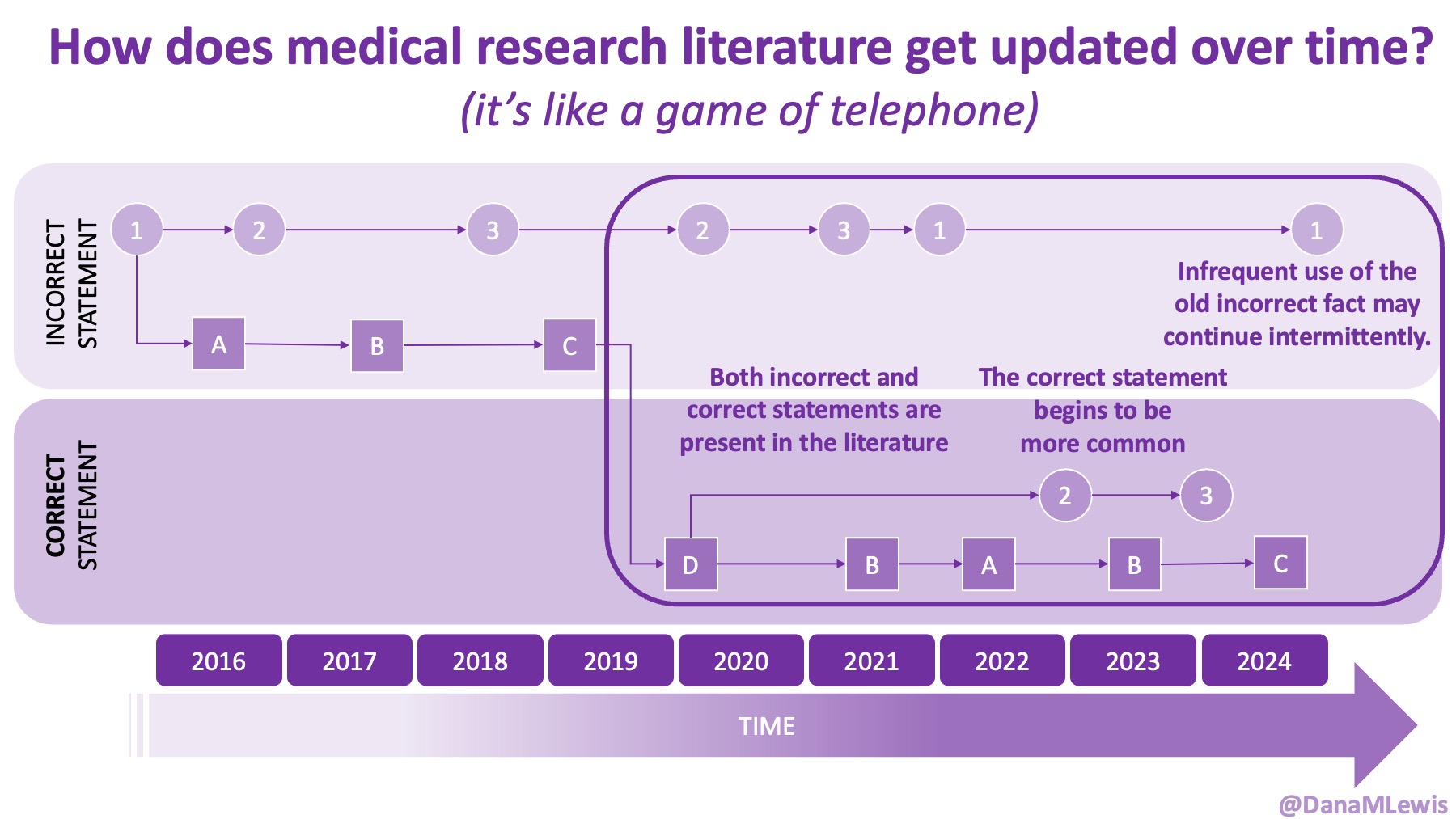
Because of these experiences, I see a lot of human AND technology errors in these spaces. I see humans make mistakes in chart notes and also in verbal comments or directions to patients. I see missed diagnoses (or wrong diagnoses) because the medical literature is like a game of telephone and the awareness of what conditions are linked to other conditions is wrong. I see LLMs make mistakes. But I see so many human mistakes in healthcare. One example from my recent personal experiences – it’s 2025 and a clinical team member asked me if I had tried cinnamon to manage my glucose levels. This was after she had done my intake and confirmed that I had type 1 diabetes. I looked at her and said no, because that would kill me. She looked surprised, then abashed when I followed that comment that I have to take insulin because I have type 1 diabetes. So I am maybe less bothered than the average person by the idea that LLMs sometimes make mistakes, say a wrong (or not quite right, even if it’s not wrong) thing, or don’t access the latest evidence base. They can, when prompted – and so can the human clinical teams.
A big point I’ve been bringing up in these talks is that we need everyone to care about the status quo of human, manual healthcare that is already riddled with errors. We need everyone to care about net risk reduction of errors and problems, not focus on additive risk.
We saw this with automated insulin delivery – people without diabetes were prone to focus on the additive risk of the technology and compare it to the zero risk they face as a person without diabetes, rather than correctly looking at the net risk assessment of how automated insulin delivery lowers the risk of living with insulin-managed diabetes even though yes, there is some additive risk. But almost a decade later, AID is now the leading therapy choice for people with insulin-managed diabetes who want it, and OS-AID is right there in the standards of care in 2026 (and has been for years!!!) with a strong evidence base.
I also talked about my experiences observing the real use of clinical scribe tools. I talked more in detail about it in this blog post, but I summarized it in my talk at Stanford, and pointed out how I was surprised to learn later – inadvertently, rather than in the consent process – that the scribe tool had access to my full medical record. It was not just transcribing the conversation and generating chart notes from it.
I also pointed out that my health system has never asked me for feedback about the tool, and that I’ve actually seen my same clinician use multiple different scribe technologies, but with no different consent process and no disclosure about chart access or any differences in privacy policy, retention timeline, etc. (Don’t pick on my clinician: she’s great. This is a point about the broader systematic failures.)
This is where I realized later people might have taken away the wrong point. This was not about me being a whiny patient, “oh they didn’t ask for my feedback! My feedback is so important!” and self-centered.
It was about flagging that these technologies do not have embedded feedback loops from ALL stakeholders, and this is more critical as we roll out more of these technologies related to healthcare.
It’s one thing to do studies and talk about user perspectives and concerns about this technology (great, and the other two presenters did an awesome job talking about their work in these spaces).
But we need to do more. We need to have pathways built in so all stakeholders – all care team members; patients; caregivers/loved ones; etc. – have pathways to talk about what is working and what is not working, from everything from errors/hallucinations in the charts to the informed consent process and how it’s actually being implemented in practice.
It matters where these pathways are implemented. The reason I haven’t forced feedback into my health system is two-fold. Yes, I have agency and the ability to give feedback when asked. Because I’ve worked at a health system before, I’m aware there’s a clinic manager and people I could go find to talk to about this. But I have bigger problems I’m dealing with (and thus limited time). And I’m not bringing it up to my clinician because we have more important things to do with our time together AND because I’m cognizant of our relationship and the power dynamics.
What do I mean? The clinician in question is new-ish to me. I’ve only been seeing her for less than two years, and I’ve been carefully building a relationship with her. Both because I’m not quite a typical patient, and because I’m not dealing with a typical situation, I’m still seeking a lot from her and her support (she’s been great, yay) to manage things while we try to figure out what’s going on. And then ditto from additional specialists, who I’m also trying to build relationships with individually, and then think about how their perceptions of me interplay with how/when they work with each other to work on my case, etc.
(Should I have to think about all this? Do I think about it too much? Maybe. But I think there’s a non-zero chance a lot of my thinking about this and how I’m coming across in my messages and appointments have played a role in the micro-successes I’ve had in the face of a rubbish and progressive health situation that no human or LLM has answers for.)
So sure, I could force my feedback into the system, but I shouldn’t have to, and I definitely shouldn’t have to be doing the risk-reward calculation on feedback directly to my clinician about this. It’s not her fault/problem/role and I don’t expect it to be: thus, what I want people to take away from my talk and this post is that I’m expecting system-level fixes and approaches that do not put more stress or risk on the patient-provider relationship.
The other thing I brought up is a question I left with everyone, which I would love to spur more discussion on. In my individual case, I have an undiagnosed situation. A team of specialists (and yes, second opinions) have not been able to diagnose/name/characterize it. I have shifted to asking providers pointedly to step away from naming and thinking broadly: what is their mental model for the mechanism of what’s going on? That’s hard to answer, because providers aren’t often thinking that way. But in this situation, when everything has been ruled out but there is CLEARLY something going on, it’s the correct thing to do.
And for the last year, the LLMs had a hard time doing it, too. Because they’re trained on the literature and the human-driven approach to make differential diagnoses, the LLMs have struggled with this. I recently began asking very specifically to work on mechanistic models. I then used that mechanistic model to frame follow up questions and discussions, to rule things in/out, to figure out where there are slight overlaps, to see where that gives us clues or evidence for/against our mechanistic model hypothesis, and to see what treatment success / failure / everything in between tells us about what is going on. Sure, a name and a diagnosis would be nice, but it’s been so relieving to have at least a hypothetical mechanistic model to use and work from. And it took specific probing (and the latest thinking/reasoning models that are now commonly available). But why am I having to do it, and why are my clinicians not doing this?
 I know some of the answers to the question of why clinicians aren’t doing this. But, the question I asked the Stanford AI+Health audience was to consider why we focus so much on informed consent for taking action, but we ignore the risks and negative outcomes that occur when not taking action.
I know some of the answers to the question of why clinicians aren’t doing this. But, the question I asked the Stanford AI+Health audience was to consider why we focus so much on informed consent for taking action, but we ignore the risks and negative outcomes that occur when not taking action.
And specifically in rare/undiagnosed diseases or edge cases of known diseases…do clinical providers have an obligation now to disclose when they are NOT using AI technology to facilitate the care they are providing?
It’s an interesting question, and one I would love to keep discussing. If you have thoughts, please share them!
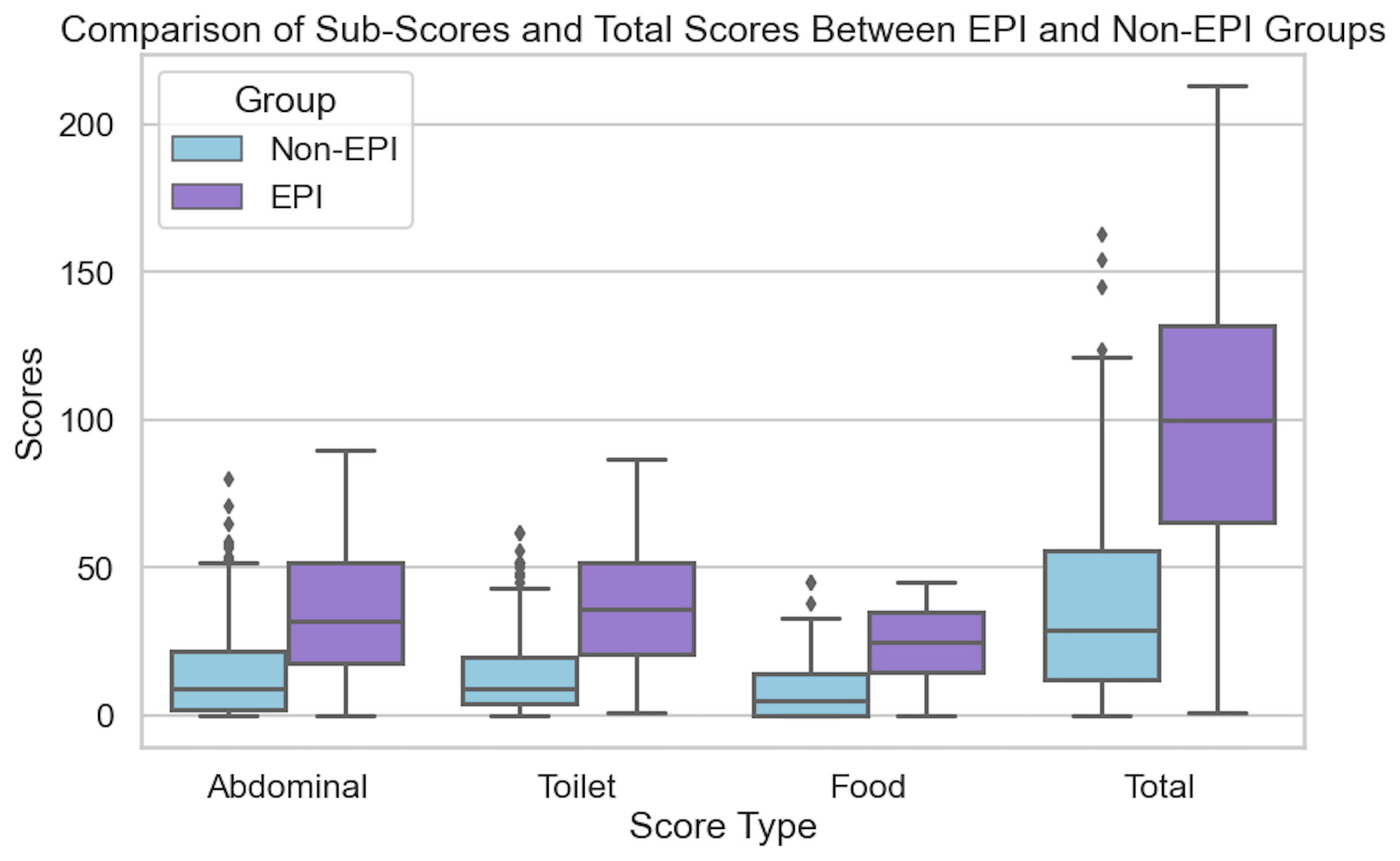
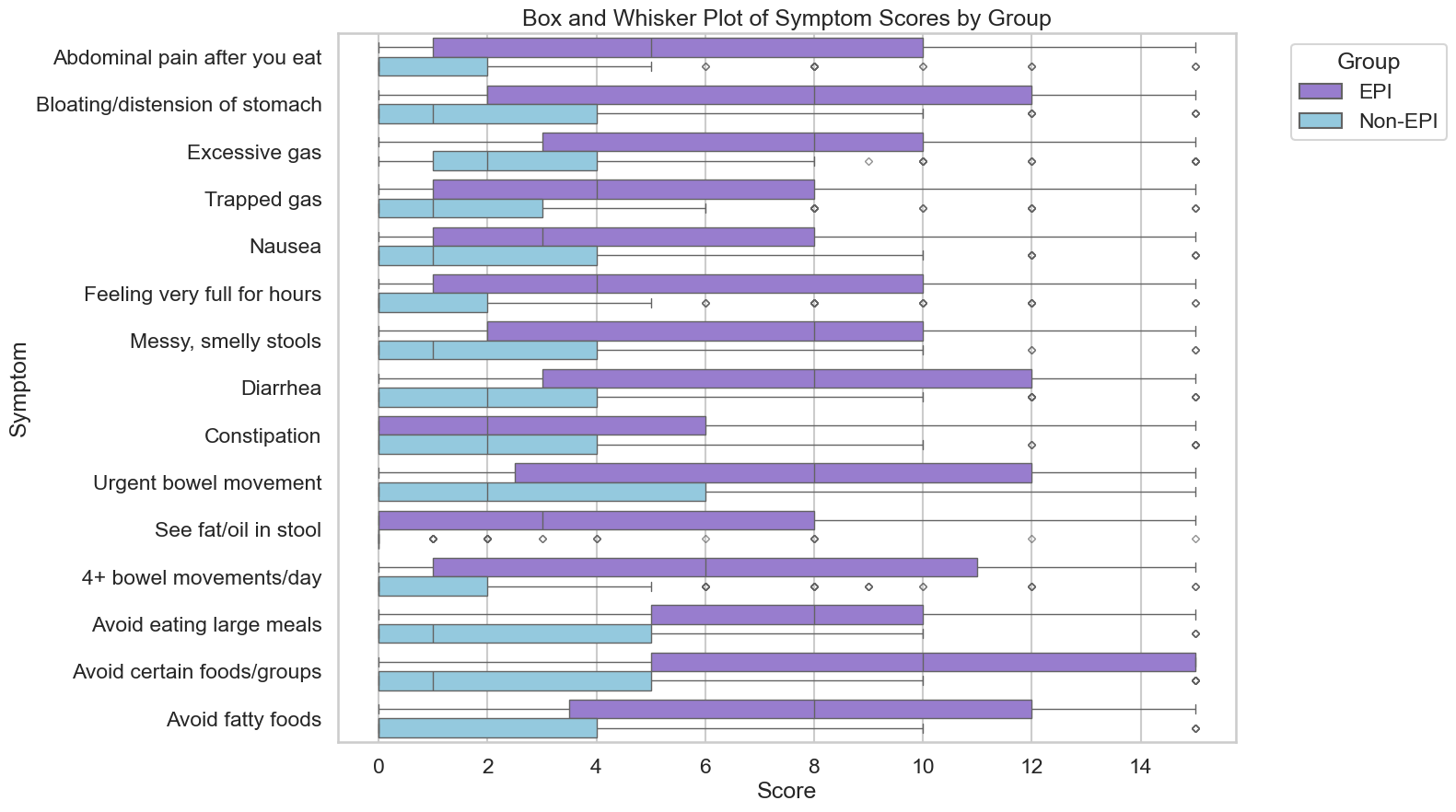
 If you have any feedback (for example, if it’s been helpful or not), you can email me any time (Dana+EPI-PEI-SS@OpenAPS.org). I’d also love to collaborate, if you’re interested in partnering on any research studies. We have some ongoing studies in different countries (US, Ireland, New Zealand, Australia) in different populations (general population; people with diabetes; people with pancreatic cancer; etc) and I’m looking forward to partnering with other researchers on additional validation studies and exploring if and how the EPI/PEI-SS can help us address some of the gaps of real-world clinical practice and life with EPI.
If you have any feedback (for example, if it’s been helpful or not), you can email me any time (Dana+EPI-PEI-SS@OpenAPS.org). I’d also love to collaborate, if you’re interested in partnering on any research studies. We have some ongoing studies in different countries (US, Ireland, New Zealand, Australia) in different populations (general population; people with diabetes; people with pancreatic cancer; etc) and I’m looking forward to partnering with other researchers on additional validation studies and exploring if and how the EPI/PEI-SS can help us address some of the gaps of real-world clinical practice and life with EPI. These are actionable, doable, practical things we can all be doing, today, and not just gnashing our teeth. The sooner we course correct with improved data availability, the better off we’ll all be in the future, whether that’s tomorrow with better clinical care or in years with AI-facilitated diagnoses, treatments, and cures.
These are actionable, doable, practical things we can all be doing, today, and not just gnashing our teeth. The sooner we course correct with improved data availability, the better off we’ll all be in the future, whether that’s tomorrow with better clinical care or in years with AI-facilitated diagnoses, treatments, and cures.  And: patients should not be punished for asking questions in order to better understand or check their understanding.
And: patients should not be punished for asking questions in order to better understand or check their understanding. 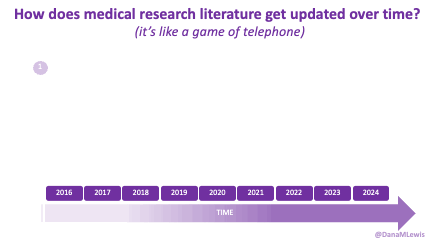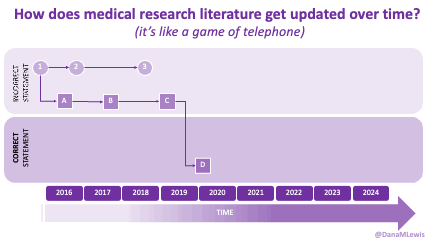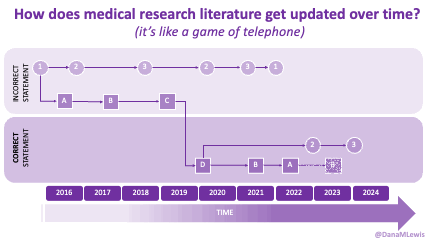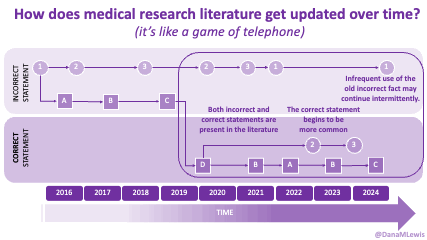
 TLDR: Instead of asking “Which model is best?”, a better question might be:
TLDR: Instead of asking “Which model is best?”, a better question might be: As an example for how I like to disseminate my articles personally, every time a journal article is published and I have access to it, I updated
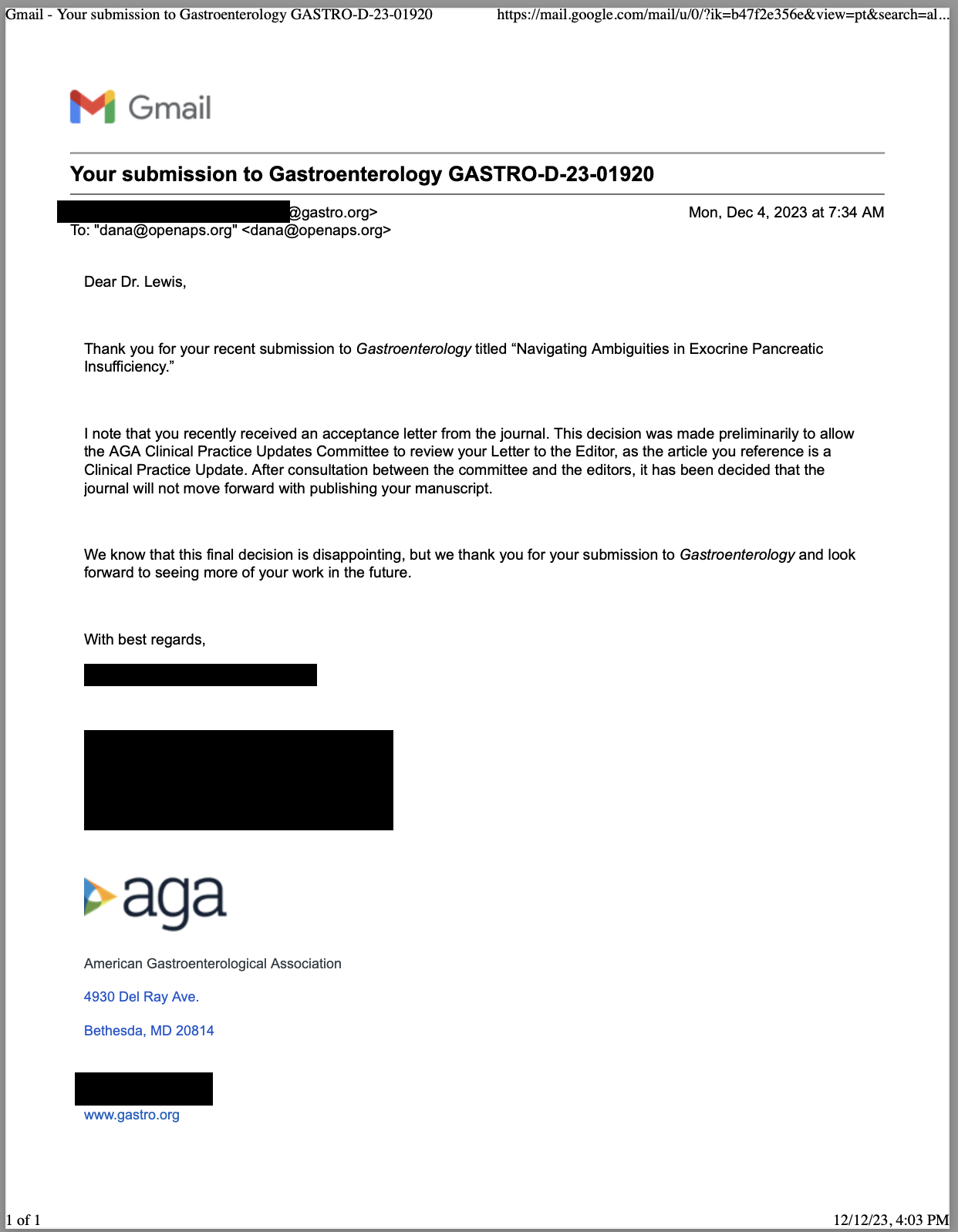
As an example for how I like to disseminate my articles personally, every time a journal article is published and I have access to it, I updated 
 How do you have people take the EPI/PEI-SS? You can pull this link up (
How do you have people take the EPI/PEI-SS? You can pull this link up (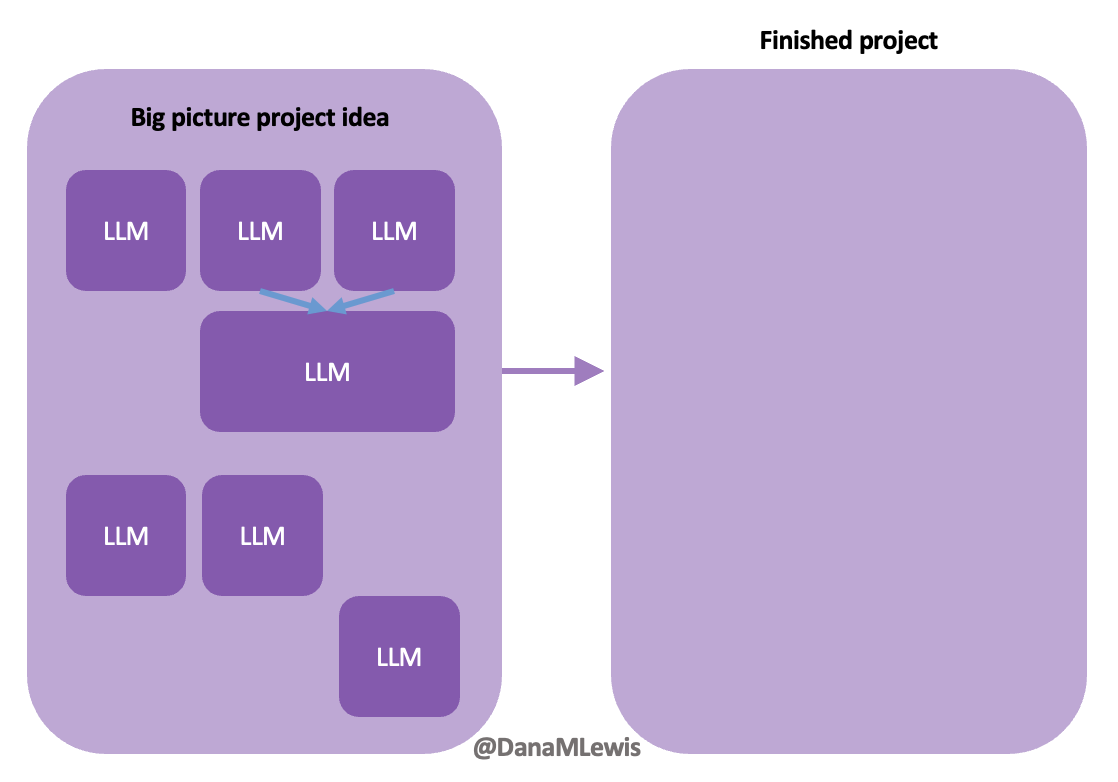 Today, I would say the same is true. It doesn’t matter – for my types of projects – if a human or an LLM “wrote” the code. What matters is: does it work as intended? Does it achieve the goal? Does it contribute to the goal of the project?
Today, I would say the same is true. It doesn’t matter – for my types of projects – if a human or an LLM “wrote” the code. What matters is: does it work as intended? Does it achieve the goal? Does it contribute to the goal of the project?



Recent Comments